Researchers at the Hong Kong University of Science and Technology (HKUST) have discovered the constructive role of a protein in driving the skeletal muscle stem cell activation to repair muscle following damage, laying the foundation for further study in the mechanisms of stem cell quiescence-to-activation transition and stem cell-based muscle regeneration.
Skeletal muscle stem cells, or satellite cells (SCs), are indispensable for repairing damaged muscle and are key targets for treating muscle diseases. In healthy uninjured muscle, these reserve stem cells lie in quiescence, a dormant state, to maintain the resident stem cell pool for future muscle repair. When muscle damage occurs, these quiescent muscle stem cells will quickly “wake up,” generating enough muscle progenitor cells to build new muscle.
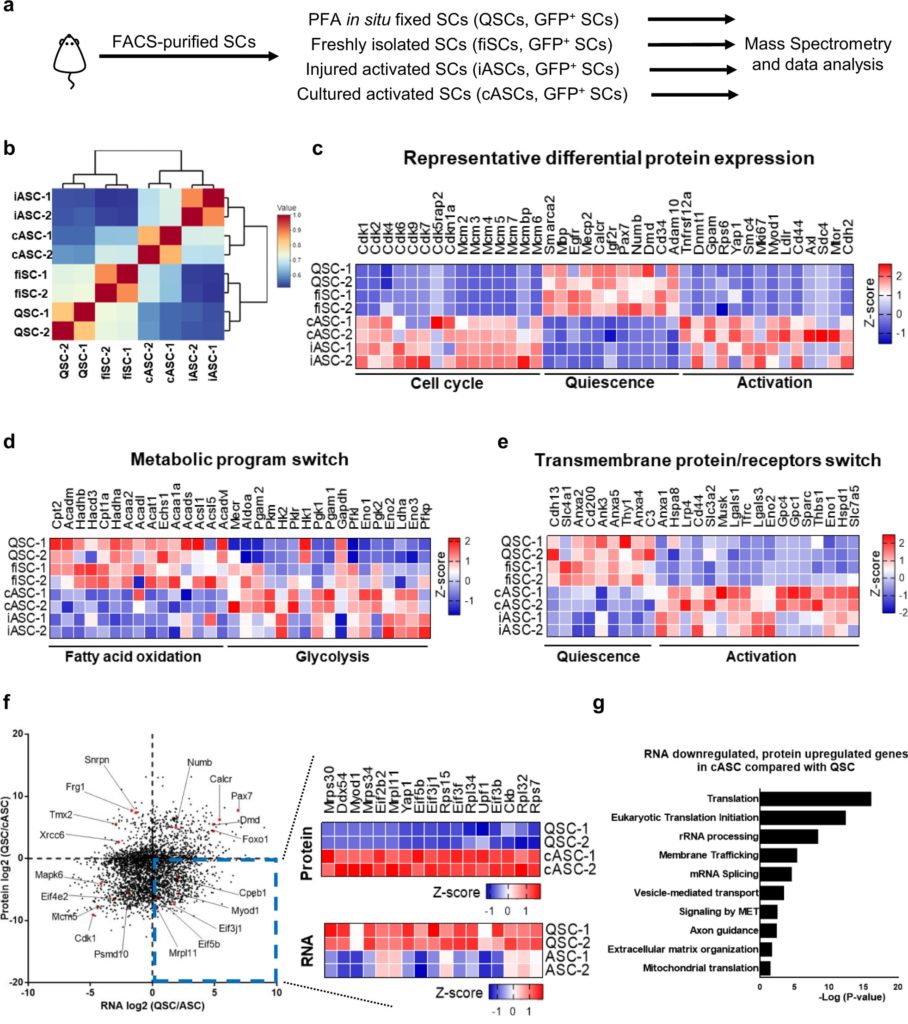
Despite being a critical step in muscle regeneration, the muscle stem cell quiescence-to-activation transition remains an elusive process, and scientists’ understanding of its mechanism and the true quiescent SC proteomics signature—the information about the entire set of proteins—has been limited.
Recently, using a whole mouse perfusion technique developed in its own laboratory to obtain the true quiescent SCs for low-input mass spectrometry analysis, a team of scientists at HKUST revealed that a regulating protein called CPEB1 is instrumental in reprogramming the translational landscape in SCs, hence driving the cells into activation and proliferation.
“In our study, we found discordance between the SC proteome and transcriptome during its activation, revealing the presence of a post-transcriptional regulation,” said Prof. Tom Cheung, lead researcher of the team and S H Ho Associate Professor of Life Science at HKUST. “Our analysis shows that levels of CPEB1 protein are low in quiescent SCs, but upregulated in activated SCs, with loss of CPEB1 delaying SC activation.”
In their subsequent RNA immunoprecipitation sequencing analysis and CPEB1-knockdown proteomic analysis, the researchers found that CPEB1 phosphorylation regulates the expression of the crucial myogenic factor MyoD—a protein involving in skeletal muscle development—by targeting some of the sequences found within the three prime untranslated region (3’UTR) of the target RNA transcript to drive SC activation.
Their findings were recently published online in the journal Nature Communications on February 17, 2022.
“It means that the manipulation of CPEB1 levels or phosphorylation can increase SC proliferation to generate enough myogenic progenitor cells for muscle repair, which could be a potential therapeutic target for muscle repair in the elderly,” noted Prof. Cheung, adding that the findings will play a fundamental role in the field as scientists continue to probe more comprehensively the mechanisms of stem cell quiescence exit and stem cell-based tissue repair.
The next step of the team’s research will involve assessing muscle regeneration in vivo in CPEB1-knockout mice to further strengthen the role of CPEB1 in SC-mediated muscle regeneration. “Furthermore, using high throughput screening, we can discover compounds that can upregulate CPEB1 protein expression to boost muscle regeneration,” Prof. Cheung said. https://medicalxpress.com/news/2022-02-scientists-reveal-mechanism-skeletal-muscle.html






Recent Comments