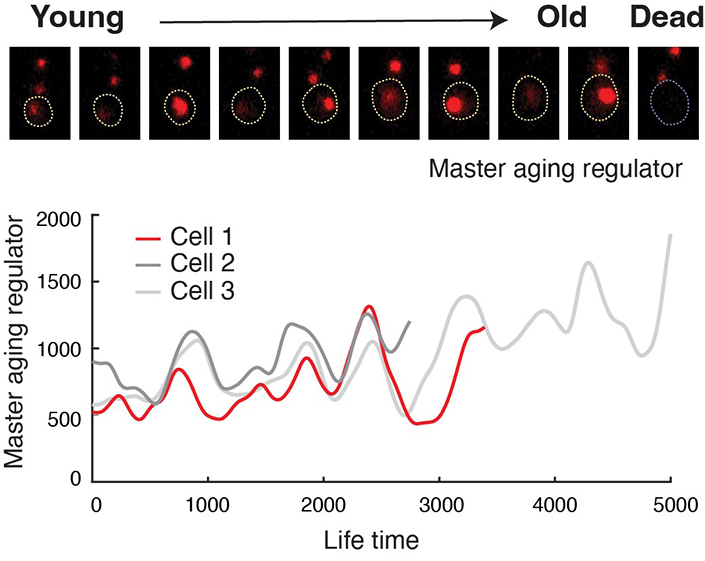
Human lifespan is related to the aging of our individual cells. Three years ago a group of University of California San Diego researchers deciphered essential mechanisms behind the aging process. After identifying two distinct directions that cells follow during aging, the researchers genetically manipulated these processes to extend the lifespan of cells.
As described in a new article published April 27, 2023, in Science, the team has now extended this research using synthetic biology to engineer a solution that keeps cells from reaching their normal levels of deterioration associated with aging.
Cells, including those of yeast, plants, animals and humans, all contain gene regu...
Read More




Recent Comments