It took 13 years and one billion dollars to sequence the human genome, an enormous scientific undertaking that launched a new era of medicine. With today’s advances in sequencing technology, that same task would have only taken about a day at a fraction of the cost. Tomorrow’s tech could whittle that down to mere seconds.
Nanopore-based DNA sequencing is a third generation technology that has the potential to further transform health care by providing rapid diagnostics of illnesses and personalizing medicine. The more efficient the method, the better. While companies have started to commercialize the technology, there are hurdles to overcome.
One nanopore method currently in use is protein-based, that is, biological. It uses membrane protein complexes that have the ability to distinguish between individual and groups of nucleotides. Unfortunately, the proteins break down with the heavy use required for sequencing—which could be millions of times for the nanopore membrane.
Solid-state nanopore sequencing, in contrast, uses synthetic materials. Two-dimensional nanomaterials such as graphene, silicon nitride, and molybdenum disulfide provide superior mechanical ability and thermal and chemical stability. But, there are still disadvantages to this method. Scientists need further investigation to better understand and characterize these different solid-state materials.
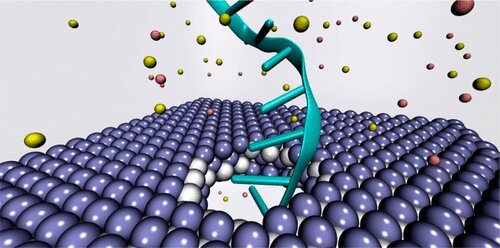
Researchers at Carnegie Mellon University became intrigued by recent developments in the synthesis of another nanomaterial, MXene. Also known as titanium carbide, it is in a class of single layer, two-dimensional inorganic compounds that are a few atoms thick. No one had previously looked to this material for use in nanopore DNA sequencing. The findings were published in the journal ACS Nano.
MXenes are notable for their properties that combine aspects of both metals and ceramics including excellent thermal and electrical conductivity, heat resistance, easy machinability, and excellent volumetric capacitance.
The researchers wanted to explore MXene as a potential membrane material for DNA detection and observe how it measured against the other nanomaterials. To investigate, they used molecular dynamics simulations to analyze its interactions with single-stranded DNA. They measured physical features such as ionic current, residence time, traces of DNA bases, physisorption, flexibility of the bases, and hydration of the nanopore.
A nanopore array can contain hundreds of pores with diameters smaller than eight nanometers. “If the nanopores are too large, all the genetic material come through the membrane mixed together,” explained Amir Barati Farimani, assistant professor of mechanical engineering. “If too small, it can’t go through at all.”
The team found that an MXene-based nanopore was able to detect different types of DNA bases with a high level of sensitivity. “We showed that MXene is an effective and promising nanomaterial for use in a nanopore-based detection platform,” said Barati Farimani.
The researchers aim to extent their work by leveraging powerful artificial intelligence (AI) algorithms to improve DNA detection by the nanopore system. DNA bases have unique features which can be used as input to train AI to improve the accuracy of DNA detection. And, AI can make use of high dimensional simulation data to learn and extract the most important features for distinguishing the DNA bases.
“The extensions to this work hold promise for vastly improving the nanopore-based detection platform and ultimately overcoming the threshold to make this technology widely applicable,” said Barati Farimani.
Other authors included Prakarsh Yadav and Zhonglin Cao, both Ph.D. students. https://phys.org/news/2021-06-nanopores-genetic-sequencing-mxene.html






Recent Comments